

Next: 6.21.3 Do's and Don'ts
Up: 6.21 GCHEM Package
Previous: 6.21.1 Introduction
Contents
FRAMEWORK
GCHEM_OPTIONS.h includes the compiler options to be used
in any experiment. For instance #define ALLOW_CFC allows
the CFC code to be run. An important compiler option is
#define GCHEM_SEPARATE_FORCING which determined
how and when the tracer forcing is applied (see discussion
on Forcing below).
There are further runtime parameters
set in data.gchem and kept in common block GCHEM.h.
These runtime options include:
 tIter0 which is the integer timestep when the tracer experiment
is initialized. If nIter0 tIter0 which is the integer timestep when the tracer experiment
is initialized. If nIter0 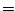 tIter0 then the tracers
are initialized to zero or from initial files. If nIter0 tIter0 then the tracers
are initialized to zero or from initial files. If nIter0 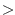 tIter0 then tracers (and previous timestep tendency terms)
are read in from a the ptracers pickup file. Note that tracers
of zeros will be carried around if nIter0
tIter0 then tracers (and previous timestep tendency terms)
are read in from a the ptracers pickup file. Note that tracers
of zeros will be carried around if nIter0 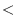 tIter0. tIter0.
 nsubtime is the integer number of extra timesteps
required by the tracer experiment. This will give a timestep
of deltaTtracer nsubtime is the integer number of extra timesteps
required by the tracer experiment. This will give a timestep
of deltaTtracer nsubtime for the dependencies
between tracers. The default is one. nsubtime for the dependencies
between tracers. The default is one.
 File names - these are several filenames than can be read in
for external fields needed in the tracer forcing - for instance
wind speed is needed in both DIC and CFC packages to calculate
the air-sea exchange of gases. Not all file names will be used
for every tracer experiment. File names - these are several filenames than can be read in
for external fields needed in the tracer forcing - for instance
wind speed is needed in both DIC and CFC packages to calculate
the air-sea exchange of gases. Not all file names will be used
for every tracer experiment.
INITIALIZATION
The values set at runtime in data.gchem are read in
using gchem_readparms.F which is called from
packages_readparms.F. This will include any external
forcing files that will be needed by the tracer experiment.
There are two routine used to initialize parameters and fields
needed by the experiment packages. These are
gchem_init_fixed.F which is called from packages_init_fixed.F, and
gchem_init_vari.F called from
packages_init_variable.F. The first should
be used to call a subroutine specific to the tracer experiment
which sets fixed parameters, the second should call a subroutine
specific to the tracer experiment
which sets (or initializes) time fields that will vary with time.
LOADING FIELDS
External forcing fields used by the tracer experiment are read
in by a subroutine (specific to the tracer experiment) called from
gchem_fields_load.F. This latter is called from forward_step.F.
FORCING
Tracer fields are advected-and-diffused by the ptracer package.
Additional changes (e.g. surface forcing or interactions
between tracers) to these fields are taken care of by the gchem
interface. For tracers that are essentially passive (e.g. CFC's)
but may have some surface boundary conditions
this can easily be done within the regular tracer timestep. In this case
gchem_calc_tendency.F is called from forward_step.F, where the
reactive (as opposed to the advective diffusive) tendencies are computed.
These tendencies, stored on the 3D field gchemTendency, are added to
the passive tracer tendencies gPtr in gchem_add_tendency.F,
which is called from ptracers_forcing.F.
For tracers with more complicated dependencies on each other,
and especially tracers which require a smaller timestep than
deltaTtracer, it will be easier to use gchem_forcing_sep.F
which is called from forward_step.F. There is a
compiler option set in GCHEM_OPTIONS.h that determines
which method is used: #define GCHEM_SEPARATE_FORCING
does the latter where tracers are forced separately from the
advection-diffusion code, and #undef GCHEM_SEPARATE_FORCING
includes the forcing in the regular timestepping.
DIAGNOSTICS
This package also also used the passive tracer routine ptracers_monitor.F
which prints out tracer statistics
as often as the model dynamic statistic diagnostics (dynsys) are written (or
as prescribed by the runtime flag PTRACERS_monitorFreq, set in data.ptracers).
There is also a placeholder for any tracer experiment
specific diagnostics to be calculated and printed to files.
This is done in gchem_diags.F. For instance the time average CO2
air-sea fluxes, and sea surface pH (among others) are written
out by dic_biotic_diags.F which is called from gchem_diags.F.



Next: 6.21.3 Do's and Don'ts
Up: 6.21 GCHEM Package
Previous: 6.21.1 Introduction
Contents
mitgcm-support@dev.mitgcm.org
| Copyright © 2002
Massachusetts Institute of Technology |
 |
|