|
|
|
|



Next: 3.13 P coordinate Global
Up: 3.12 Global Ocean MITgcm
Previous: 3.12.2 Discrete Numerical Configuration
Contents
Subsections
3.12.3 Experiment Configuration
The model configuration for this experiment resides under the
directory tutorial_global_oce_latlon/. The experiment files
- input/data
- input/data.pkg
- input/eedata,
- input/trenberth_taux.bin,
- input/trenberth_tauy.bin,
- input/lev_s.bin,
- input/lev_t.bin,
- input/lev_sss.bin,
- input/lev_sst.bin,
- input/bathymetry.bin,
- code/SIZE.h.
contain the code customizations and parameter settings for these
experiments. Below we describe the customizations
to these files associated with this experiment.
Figures (3.5-3.10)
show the relaxation temperature (
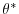 ) and salinity (
) and salinity (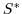 )
fields, the wind stress components (
)
fields, the wind stress components (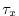 and
and 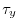 ), the heat flux (
), the heat flux (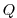 )
and the net fresh water flux (
)
and the net fresh water flux (
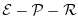 ) used
in equations
(3.31-3.34).
The figures also indicate the lateral extent and coastline used in the
experiment. Figure (-- missing figure -- ) shows the depth contours of the model domain.
) used
in equations
(3.31-3.34).
The figures also indicate the lateral extent and coastline used in the
experiment. Figure (-- missing figure -- ) shows the depth contours of the model domain.
This file, reproduced completely below, specifies the main parameters
for the experiment. The parameters that are significant for this configuration
are
- Lines 7-8
tRef= 15*20.,
sRef= 15*35.,
set reference values for potential
temperature and salinity at each model level in units of
 and
and
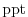 . The entries are ordered from surface to depth.
Density is calculated from anomalies at each level evaluated
with respect to the reference values set here.
. The entries are ordered from surface to depth.
Density is calculated from anomalies at each level evaluated
with respect to the reference values set here.

- Line 9,
viscAr=1.E-3,
this line sets the vertical Laplacian dissipation coefficient to
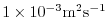 . Boundary conditions
for this operator are specified later.
. Boundary conditions
for this operator are specified later.

- Line 10,
viscAh=5.E5,
this line sets the horizontal Laplacian frictional dissipation coefficient to
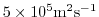 . Boundary conditions
for this operator are specified later.
. Boundary conditions
for this operator are specified later.
- Lines 11 and 13,
diffKhT=0.,
diffKhS=0.,
set the horizontal diffusion coefficient for temperature and salinity
to 0, since package GMREDI is used.
- Lines 12 and 14,
diffKrT=3.E-5,
diffKrS=3.E-5,
set the vertical diffusion coefficient for temperature and salinity
to
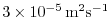 . The boundary
condition on this operator is
. The boundary
condition on this operator is
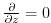 at both
the upper and lower boundaries.
at both
the upper and lower boundaries.
- Lines 15-17
rhonil=1035.,
rhoConstFresh=1000.,
eosType = 'JMD95Z',
set the reference densities for sea water and fresh water, and selects
the equation of state [Jackett and McDougall, 1995]
- Lines 18-19,
ivdc_kappa=100.,
implicitDiffusion=.TRUE.,
specify an ``implicit diffusion'' scheme with increased vertical
diffusivity of 100 m /s in case of instable stratification.
/s in case of instable stratification.

- ...
- Line 28,
readBinaryPrec=32,
Sets format for reading binary input datasets holding model fields to
use 32-bit representation for floating-point numbers.
- Line 33,
cg2dMaxIters=500,
Sets maximum number of iterations the two-dimensional, conjugate
gradient solver will use, irrespective of convergence
criteria being met.

- Line 34,
cg2dTargetResidual=1.E-13,
Sets the tolerance which the two-dimensional, conjugate
gradient solver will use to test for convergence in equation
2.20 to
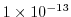 .
Solver will iterate until tolerance falls below this value or until the
maximum number of solver iterations is reached.
.
Solver will iterate until tolerance falls below this value or until the
maximum number of solver iterations is reached.

- Line 39,
nIter0=0,
Sets the starting time for the model internal time counter.
When set to non-zero this option implicitly requests a
checkpoint file be read for initial state.
By default the checkpoint file is named according to
the integer number of time step value nIter0.
The internal time counter works in seconds. Alternatively,
startTime can be set.
- Line 40,
nTimeSteps=20,
Sets the time step number at which this simulation will terminate.
At the end of a simulation a checkpoint file is automatically
written so that a numerical experiment can consist of multiple
stages. Alternatively endTime can be set.
- Line 44,
deltaTmom=1800.,
Sets the timestep
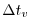 used in the momentum equations to
used in the momentum equations to
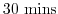 .
See section 2.2.
.
See section 2.2.

- Line 45,
tauCD=321428.,
Sets the D-grid to C-grid coupling time scale 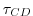 used in the momentum equations.
used in the momentum equations.

- Lines 46-48,
deltaTtracer=86400.,
deltaTClock = 86400.,
deltaTfreesurf= 86400.,
Sets the default timestep,
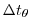 , for tracer equations
and implicit free surface equations to
, for tracer equations
and implicit free surface equations to
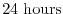 .
See section 2.2.
.
See section 2.2.

- Line 76,
bathyFile='bathymetry.bin'
This line specifies the name of the file from which the domain
bathymetry is read. This file is a two-dimensional (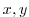 ) map of
depths. This file is assumed to contain 32-bit binary numbers
giving the depth of the model at each grid cell, ordered with the x
coordinate varying fastest. The points are ordered from low coordinate
to high coordinate for both axes. The units and orientation of the
depths in this file are the same as used in the MITgcm code. In this
experiment, a depth of
) map of
depths. This file is assumed to contain 32-bit binary numbers
giving the depth of the model at each grid cell, ordered with the x
coordinate varying fastest. The points are ordered from low coordinate
to high coordinate for both axes. The units and orientation of the
depths in this file are the same as used in the MITgcm code. In this
experiment, a depth of 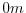 indicates a solid wall and a depth
of
indicates a solid wall and a depth
of 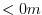 indicates open ocean.
indicates open ocean.
- Lines 79-80,
zonalWindFile='trenberth_taux.bin'
meridWindFile='trenberth_tauy.bin'
These lines specify the names of the files from which the x- and y-
direction surface wind stress is read. These files are also
three-dimensional (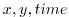 ) maps and are enumerated and formatted
in the same manner as the bathymetry file.
) maps and are enumerated and formatted
in the same manner as the bathymetry file.
other lines in the file input/data are standard values
that are described in the MITgcm Getting Started and MITgcm Parameters
notes.
1 # ====================
2 # | Model parameters |
3 # ====================
4 #
5 # Continuous equation parameters
6 &PARM01
7 tRef = 15*20.,
8 sRef = 15*35.,
9 viscAr=1.E-3,
10 viscAh=5.E5,
11 diffKhT=0.,
12 diffKrT=3.E-5,
13 diffKhS=0.,
14 diffKrS=3.E-5,
15 rhonil=1035.,
16 rhoConstFresh=1000.,
17 eosType = 'JMD95Z',
18 ivdc_kappa=100.,
19 implicitDiffusion=.TRUE.,
20 allowFreezing=.TRUE.,
21 exactConserv=.TRUE.,
22 useRealFreshWaterFlux=.TRUE.,
23 useCDscheme=.TRUE.,
24 # turn on looped cells
25 hFacMin=.05,
26 hFacMindr=50.,
27 # set precision of data files
28 readBinaryPrec=32,
29 &
30
31 # Elliptic solver parameters
32 &PARM02
33 cg2dMaxIters=500,
34 cg2dTargetResidual=1.E-13,
35 &
36
37 # Time stepping parameters
38 &PARM03
39 nIter0= 0,
40 nTimeSteps = 20,
41 # 100 years of integration will yield a reasonable flow field
42 # startTime = 0.,
43 # endTime = 3110400000.,
44 deltaTmom = 1800.,
45 tauCD = 321428.,
46 deltaTtracer= 86400.,
47 deltaTClock = 86400.,
48 deltaTfreesurf= 86400.,
49 abEps = 0.1,
50 pChkptFreq= 1728000.,
51 dumpFreq= 864000.,
52 taveFreq= 864000.,
53 monitorFreq=1.,
54 # 2 months restoring timescale for temperature
55 tauThetaClimRelax= 5184000.,
56 # 6 months restoring timescale for salinity
57 tauSaltClimRelax = 15552000.,
58 periodicExternalForcing=.TRUE.,
59 externForcingPeriod=2592000.,
60 externForcingCycle=31104000.,
61 &
62
63 # Gridding parameters
64 &PARM04
65 usingSphericalPolarGrid=.TRUE.,
66 delR= 50., 70., 100., 140., 190.,
67 240., 290., 340., 390., 440.,
68 490., 540., 590., 640., 690.,
69 ygOrigin=-80.,
70 dySpacing=4.,
71 dxSpacing=4.,
72 &
73
74 # Input datasets
75 &PARM05
76 bathyFile= 'bathymetry.bin',
77 hydrogThetaFile='lev_t.bin',
78 hydrogSaltFile= 'lev_s.bin',
79 zonalWindFile= 'trenberth_taux.bin',
80 meridWindFile= 'trenberth_tauy.bin',
81 thetaClimFile= 'lev_sst.bin',
82 saltClimFile= 'lev_sss.bin',
83 surfQFile= 'ncep_qnet.bin',
84 the_run_name= 'global_oce_latlon',
85 # fresh water flux is turned on, comment next line to it turn off
86 # (maybe better with surface salinity restoring)
87 EmPmRFile= 'ncep_emp.bin',
88 &
This file uses standard default values and does not contain
customisations for this experiment.
This file uses standard default values and does not contain
customisations for this experiment.
The input/trenberth_taux.bin and input/trenberth_tauy.bin files specify a three-dimensional
(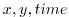 ) map of wind stress,
) map of wind stress,
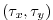 , values
[Trenberth et al., 1990]. The units used are
, values
[Trenberth et al., 1990]. The units used are 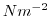 .
.
The input/bathymetry.bin file specifies a two-dimensional
(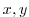 ) map of depth values. For this experiment values range
between 0
and
) map of depth values. For this experiment values range
between 0
and
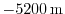 , and have been derived from
ETOPO5. The file contains a raw binary stream of data that is
enumerated in the same way as standard MITgcm two-dimensional,
horizontal arrays.
, and have been derived from
ETOPO5. The file contains a raw binary stream of data that is
enumerated in the same way as standard MITgcm two-dimensional,
horizontal arrays.
Four lines are customized in this file for the current experiment
- Line 40,
sNx=45,
this line sets the number of grid points of each tile (or sub-domain)
along the x-coordinate axis.
- Line 41,
sNy=40,
this line sets the number of grid points of each tile (or sub-domain)
along the y-coordinate axis.
- Lines 46 and 44,
nPx=1,
nSx=2,
theses lines set, respectively, the number of processes and
the number of tiles per process along the x-coordinate axis.
Therefore, the total number of grid points along the x-coordinate axis
corresponding to the full domain extent is
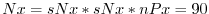
- Line 50,
Nr=15
this line sets the vertical domain extent in grid points.
1 C $Header: /u/gcmpack/MITgcm/verification/tutorial_global_oce_latlon/code/SIZE.h,v 1.1 2006/07/14 20:35:22 jmc Exp $
2 C $Name: $
3
4 C
5 C /==========================================================\
6 C | SIZE.h Declare size of underlying computational grid. |
7 C |==========================================================|
8 C | The design here support a three-dimensional model grid |
9 C | with indices I,J and K. The three-dimensional domain |
10 C | is comprised of nPx*nSx blocks of size sNx along one axis|
11 C | nPy*nSy blocks of size sNy along another axis and one |
12 C | block of size Nz along the final axis. |
13 C | Blocks have overlap regions of size OLx and OLy along the|
14 C | dimensions that are subdivided. |
15 C \==========================================================/
16 C Voodoo numbers controlling data layout.
17 C sNx - No. X points in sub-grid.
18 C sNy - No. Y points in sub-grid.
19 C OLx - Overlap extent in X.
20 C OLy - Overlat extent in Y.
21 C nSx - No. sub-grids in X.
22 C nSy - No. sub-grids in Y.
23 C nPx - No. of processes to use in X.
24 C nPy - No. of processes to use in Y.
25 C Nx - No. points in X for the total domain.
26 C Ny - No. points in Y for the total domain.
27 C Nr - No. points in Z for full process domain.
28 INTEGER sNx
29 INTEGER sNy
30 INTEGER OLx
31 INTEGER OLy
32 INTEGER nSx
33 INTEGER nSy
34 INTEGER nPx
35 INTEGER nPy
36 INTEGER Nx
37 INTEGER Ny
38 INTEGER Nr
39 PARAMETER (
40 & sNx = 45,
41 & sNy = 40,
42 & OLx = 2,
43 & OLy = 2,
44 & nSx = 2,
45 & nSy = 1,
46 & nPx = 1,
47 & nPy = 1,
48 & Nx = sNx*nSx*nPx,
49 & Ny = sNy*nSy*nPy,
50 & Nr = 15)
51
52 C MAX_OLX - Set to the maximum overlap region size of any array
53 C MAX_OLY that will be exchanged. Controls the sizing of exch
54 C routine buufers.
55 INTEGER MAX_OLX
56 INTEGER MAX_OLY
57 PARAMETER ( MAX_OLX = OLx,
58 & MAX_OLY = OLy )
59



Next: 3.13 P coordinate Global
Up: 3.12 Global Ocean MITgcm
Previous: 3.12.2 Discrete Numerical Configuration
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2018-01-23 |
 |
|







