|
|
|
|



Next: 3.20.4 Running The Example
Up: 3.20 Offline Example
Previous: 3.20.2 Time-stepping of tracers
Contents
Subsections
3.20.3 Code Configuration
The model configuration for this experiment resides under the
directory verification/tutorial_offline. The experiment files
- input/data
- input/data.off
- input/data.pkg
- input/data.ptracers
- input/eedata
- input/packages.conf
- code/PTRACERS_SIZE.h
- code/SIZE.h.
contain the code customisations and parameter settings
required to run the example. In addition the following binary data
files are required:
- input/depth_g77.bin
- input/tracer1_.bin
- input/tracer2_.bin
- input/input_off/uVeltave.0000000001-12.data
- input/input_off/vVeltave.0000000001-12.data
- input/input_off/wVeltave.0000000001-12.data
- input/input_off/Convtave.0000000001-12.data
This file, reproduced completely below, specifies the main
parameters for the experiment.
nIter0 and nTimesteps control the start time and the length
of the run (in timesteps). If nIter0 is non-zero the model will
require appropriate pickup files to be present in the run directory.
Where nIter0 is zero, as here, the model makes a fresh start. In this
case the model has been prescribed to run for 720 timesteps or 1 year.
- Line 20
deltaTtracer= 43200.0,
deltaTtracer is the tracer timestep in seconds, in this
case, 12 hours (43200s = 12 hours). Note that deltatTracer must be
specified in input/data as well as specifying deltaToffline in
input/data.off.
- Line 21
deltaTClock= 43200.0,
When using the MITgcm in offline mode deltaTClock (an
internal model counter) should be made equal to the value assigned to
deltatTtracer.
periodicExternalForcing is a flag telling the model whether
to cyclically re-use forcing data where there is external forcing (see
`A More Complcated Example', below). Where there is no external
forcing, as here, but where there is to be cyclic re-use of the
offline flow and mixing fields, periodicExternalForcing must be
assigned the value .TRUE.
externForcingPeriod specifies the period of the external
forcing data in seconds. In the absence of external forcing, as in
this example, it must be made equal to the value of
externForcingPeriod in input/data.off, in this case, monthly
(2592000s = 1 month).
externForcingCycle specifies the duration of the external
forcing data cycle in seconds. In the absence of external forcing, as
in this example, it must be made equal to the value of
externForcingCycle in input/data.off, in this case, the cycle is
one year(31104000s = 1 year).
This line requests that the simulation be performed in a
spherical polar coordinate system. It affects the interpretation of
grid input parameters and causes the grid generation routines to
initialize an internal grid based on spherical polar geometry.
- Line 36
delR= 50., 70., 100., 140., 190.,
240., 290., 340., 390., 440.,
490., 540., 590., 640., 690.,
This line sets the vertical grid spacing between each
z-coordinate line in the discrete grid. Here the total model depth is
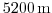 .
.
This line sets the southern boundary of the modeled domain
to
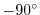 latitude N (
latitude N (
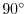 S). This value affects both
the generation of the
locally orthogonal grid that the model uses internally and affects the
initialization of the coriolis force. Note - it is not required to
set a longitude boundary, since the absolute longitude does not alter
the kernel equation discretisation.
S). This value affects both
the generation of the
locally orthogonal grid that the model uses internally and affects the
initialization of the coriolis force. Note - it is not required to
set a longitude boundary, since the absolute longitude does not alter
the kernel equation discretisation.
- Line 40
dxSpacing=2.8125,
This line sets the horizontal grid spacing between each
y-coordinate line in the discrete grid to
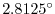 in
longitude.
in
longitude.
- Line 41
dySpacing=2.8125,
This line sets the vertical grid spacing between each
x-coordinate line in the discrete grid to
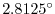 in
latitude.
in
latitude.
This line specifies the name of the file, in this case depth_g77.bin, from which the domain bathymetry is read. This file
contains a two-dimensional (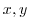 ) map of (assumed 64-bit) binary
numbers giving the depth of the model at each grid cell, ordered with
the x coordinate varying fastest. The points are ordered from low
coordinate to high coordinate for both axes. The units and orientation
of the depths in this file are the same as used in the MITgcm code. In
this experiment, a depth of
) map of (assumed 64-bit) binary
numbers giving the depth of the model at each grid cell, ordered with
the x coordinate varying fastest. The points are ordered from low
coordinate to high coordinate for both axes. The units and orientation
of the depths in this file are the same as used in the MITgcm code. In
this experiment, a depth of 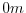 indicates land.
indicates land.
1 # ====================
2 # | Model parameters |
3 # ====================
4 #
5 # Continuous equation parameters
6 &PARM01
7 &
8 #
9 # Elliptic solver parameters
10 &PARM02
11 cg2dMaxIters=1000,
12 cg2dTargetResidual=1.E-13,
13 &
14 #
15
16 # Time stepping parameters
17 &PARM03
18 nIter0 = 0,
19 nTimeSteps = 720,
20 deltaTtracer= 43200.0,
21 deltaTClock = 43200.0,
22 pChkptFreq=31104000.,
23 chkptFreq= 31104000.,
24 dumpFreq= 2592000.,
25 taveFreq= 311040000.,
26 monitorFreq= 1.,
27 periodicExternalForcing=.TRUE.,
28 externForcingPeriod=2592000.,
29 externForcingCycle=31104000.,
30 &
31 #
32 # Gridding parameters
33 &PARM04
34 usingCartesianGrid=.FALSE.,
35 usingSphericalPolarGrid=.TRUE.,
36 delR= 50., 70., 100., 140., 190.,
37 240., 290., 340., 390., 440.,
38 490., 540., 590., 640., 690.,
39 ygOrigin=-90.,
40 dxSpacing=2.8125,
41 dySpacing=2.8125,
42 &
43 #
44 # Input datasets
45 &PARM05
46 bathyFile= 'depth_g77.bin',
47 &
input/data.off provides the MITgcm offline package
with package specific parameters. input/data.off specifies the
location (relative to the run directory) and prefix of files
describing the flow field (UvelFile, VvelFile, WvelFile) and the
corresponding convective mixing coefficients (ConvFile) which together
prescribe the three dimensional, time varying dynamic system within
which the offline model will advect the tracer.
- Lines 2 to 5
UvelFile= '../input/input_off/uVeltave',
VvelFile= '../input/input_off/vVeltave',
WvelFile= '../input/input_off/wVeltave',
ConvFile= '../input/input_off/Convtave',
In the example the offline data is located in the
sub-directory input/input_off. In this directory are fields
describing the velocity and convective mixing histories of a prior
forward integration of the MITgcm required for the offline package and
identified in input/data_off. Based on the values of deltatToffline, offlineForcingPeriod and offlineForcingCycle specified in input/data.off, since offlineForcingCycle corresponds to 12 forcing periods offlineForcingPeriod and since offlineIter0 is zero, there needs to
be 12 uVeltave, 12 vVeltave, 12 wVeltave and 12 Convtave files each
having a 10 digit sequence identifier between 0000000001 to
0000000012, that is, a total of 48 files.
offlineIter0, here specified to be 0 timesteps, corresponds
to the timestep at which the tracer model is initialised. Note that
offlineIter0 and nIter0 (set in input/data) need not be the
same.
- Line 10
deltaToffline=43200.,
deltatToffline sets the timestep associated with the offline
model data in seconds, here 12 hours (43200s = 12 hours).
offlineForcingPeriod sets the forcing period associated with
the offline model data in seconds.
offlineForcingCycle sets the forcing cycle length associated
with the offline model data in seconds. In this example the offline
forcing cycle is 6 days, or 12 offline forcing periods. Together
deltatToffline, offlineForcingPeriod and offlineForcingCycle determine
the value of the 10 digit sequencing tag the model expects files in
input/input_off to have.
1 &OFFLINE_PARM01
2 UvelFile= 'input_off/uVeltave',
3 VvelFile= 'input_off/vVeltave',
4 WvelFile= 'input_off/wVeltave',
5 ConvFile= 'input_off/Convtave',
6 &end
7 &OFFLINE_PARM02
8 offlineIter0=0,
9 deltaToffline=43200.,
10 offlineForcingPeriod=43200.,
11 offlineForcingCycle=518400.,
12 &end
File input/data.pkg, reproduced completely below,
specifies which MITgcm packages ( /MITgcm/pkg) are to be used.
- Line 3
usePTRACERS=.TRUE.,
usePTRACERS is a flag invoking the ptracers package which is
responsible for the advection of the tracer within the model.
1 # Packages
2 &PACKAGES
3 usePTRACERS=.TRUE.,
4 &
File input/data.ptracers, reproduced completely below,
provides the MITgcm ptracers package with package specific parameters,
prescribing the nature of the the tracer/tracers as well as the
variables associated with their advection.
- Line 2
PTRACERS_numInUse=2,
PTRACERS_numInUse tells the model how many separate tracers
are to be advected, in this case 2. Note: The value of
PTRACERS_numInUse must agree with the value specified in code/PTRACERS_SIZE.h - see code/PTRACERS_SIZE.h below.
- Line 3
PTRACERS_Iter0= 0,
PTRACERS_Iter0 specifies the iteration at which the tracer
is to be introduced. In this case the tracer is initialised at the
start of the simulation. i.e. PTRACERS_Iter0 = PTRACERS_nIter0.
PTRACERS_advScheme(n) identifies which advection scheme
will be used for tracer n, where n is the number of the tracer up to
PTRACERS_numInUse. See section 2.18, 'Comparison of advection
schemes', to identify the numerical codes used to specify different
advection schemes (e.g. centered 2nd order, 3rd order upwind) as well
as details of each.
- Lines 6 and 11
PTRACERS_diffKh(1)=1.E3,
PTRACERS_diffKh(n) is the horizontal diffusion coefficient
for tracer n, where n is the number of the tracer up to
PTRACERS_numInUse.
PTRACERS_diffKr(n) is the vertical diffusion coefficient
for tracer n, where n is the number of the tracer up to
PTRACERS_numInUse.
PTRACERS_initialFile(n) identifies the initial tracer field
to be associated with tracer n, where n is the number of the tracer up
to PTRACERS_numInUse. In this example file input/tracer1.bin
contains localised tracer, input/tracer2.bin contains an
arbitrary global distribution. Included Matlab script input/makeinitialtracer.m provides a template for generating or
manipulating initial tracer fields.
1 &PTRACERS_PARM01
2 PTRACERS_numInUse=2,
3 PTRACERS_Iter0= 0,
4 # tracer 1
5 PTRACERS_advScheme(1)=77,
6 PTRACERS_diffKh(1)=1.E3,
7 PTRACERS_diffKr(1)=5.E-5,
8 PTRACERS_initialFile(1)='tracer1.bin',
9 # tracer 2
10 PTRACERS_advScheme(2)=77,
11 PTRACERS_diffKh(2)=1.E3,
12 PTRACERS_diffKr(2)=5.E-5,
13 PTRACERS_initialFile(2)='tracer2.bin',
14 &
Note input/data.ptracers requires a set of entries for
each tracer.
This file uses standard default values and does not contain
customisations for this experiment.
The following code changes are required to run this exaperiment.
This file is used to invoke the model components required
for a particuylar implementation of the MITgcm. In this case the code/packages.conf contains the component names:
ptracers
generic_advdiff
mdsio
mom_fluxform
mom_vecinv
timeave
rw
monitor
offline
This line sets the parameters PTRACERS_num (the number of
tracers to be integrated) to 2 (in agreement with input/data.ptracers).
Two lines are customized in this file for the current
experiment
- Line 39,
sNx=128,
this line sets
the lateral domain extent in grid points for the
axis aligned with the x-coordinate.
- Line 40,
sNy=64,
this line sets
the lateral domain extent in grid points for the
axis aligned with the y-coordinate.
- Line 49,
Nr=15,
this line sets
the vertical domain extent in grid points.



Next: 3.20.4 Running The Example
Up: 3.20 Offline Example
Previous: 3.20.2 Time-stepping of tracers
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2018-01-23 |
 |
|







