|
|
|
|



Next: 4. Software Architecture
Up: 3.21 A Rotating Tank
Previous: 3.21.3 Discrete Numerical Configuration
Contents
Subsections
3.21.4 Code Configuration
The model configuration for this experiment resides under the
directory verification/rotatingi_tank/. The experiment files
- input/data
- input/data.pkg
- input/eedata,
- input/bathyPol.bin,
- input/thetaPol.bin,
- code/CPP_EEOPTIONS.h
- code/CPP_OPTIONS.h,
- code/SIZE.h.
contain the code customizations and parameter settings for this
experiments. Below we describe the customizations
to these files associated with this experiment.
This file, reproduced completely below, specifies the main parameters
for the experiment. The parameters that are significant for this configuration
are
- Lines 9-10,
viscAh=5.0E-6,
viscAz=5.0E-6,
These lines set the Laplacian friction coefficient in the horizontal
and vertical, respectively. Note that they are several orders of
magnitude smaller than the other examples due to the small scale of
this example.
- Lines 13-16,
diffKhT=2.5E-6,
diffKzT=2.5E-6,
diffKhS=1.0E-6,
diffKzS=1.0E-6,
These lines set horizontal and vertical diffusion coefficients for
temperature and salinity. Similarly to the friction coefficients, the
values are a couple of orders of magnitude less than most
configurations.
- Line 17,
f0=0.5 ,
this line sets the
coriolis term, and represents a tank spinning at about 2.4 rpm.
- Lines 23 and 24
rigidLid=.TRUE.,
implicitFreeSurface=.FALSE.,
These lines activate the rigid lid formulation of the surface
pressure inverter and suppress the implicit free surface form
of the pressure inverter.
- Line 40,
nIter=0,
This line indicates that the experiment should start from 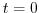 and
implicitly suppresses searching for checkpoint files associated with
restarting an numerical integration from a previously saved state.
Instead, the file thetaPol.bin will be loaded to initialized the
temperature fields as indicated below, and other variables will be
initialized to their defaults.
and
implicitly suppresses searching for checkpoint files associated with
restarting an numerical integration from a previously saved state.
Instead, the file thetaPol.bin will be loaded to initialized the
temperature fields as indicated below, and other variables will be
initialized to their defaults.
- Line 43,
deltaT=0.1,
This line sets the integration timestep to 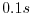 . This is an
unsually small value among the examples due to the small physical
scale of the experiment. Using the ensemble Kalman filter to produce
input fields can necessitate even shorter timesteps.
. This is an
unsually small value among the examples due to the small physical
scale of the experiment. Using the ensemble Kalman filter to produce
input fields can necessitate even shorter timesteps.
- Line 56,
usingCylindricalGrid=.TRUE.,
This line requests that the simulation be performed in a
cylindrical coordinate system.
- Line 57,
dXspacing=3,
This line sets the azimuthal grid spacing between each 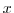 -coordinate line
in the discrete grid. The syntax indicates that the discrete grid
should be comprised of
-coordinate line
in the discrete grid. The syntax indicates that the discrete grid
should be comprised of 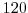 grid lines each separated by
grid lines each separated by 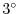 .
.
- Line 58,
dYspacing=0.01,
This line sets the radial cylindrical grid spacing between each
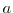 -coordinate line in the discrete grid to
-coordinate line in the discrete grid to 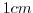 .
.
- Line 59,
delZ=29*0.005,
This line sets the vertical grid spacing between each of 29
z-coordinate lines in the discrete grid to 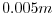 (
(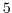 mm).
mm).
- Line 64,
bathyFile='bathyPol.bin',
This line specifies the name of the file from which the domain
``bathymetry'' (tank depth) is read. This file is a two-dimensional
(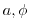 ) map of
depths. This file is assumed to contain 64-bit binary numbers
giving the depth of the model at each grid cell, ordered with the
) map of
depths. This file is assumed to contain 64-bit binary numbers
giving the depth of the model at each grid cell, ordered with the 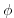 coordinate varying fastest. The points are ordered from low coordinate
to high coordinate for both axes. The units and orientation of the
depths in this file are the same as used in the MITgcm code. In this
experiment, a depth of
coordinate varying fastest. The points are ordered from low coordinate
to high coordinate for both axes. The units and orientation of the
depths in this file are the same as used in the MITgcm code. In this
experiment, a depth of 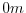 indicates an area outside of the tank
and a depth
f
indicates an area outside of the tank
and a depth
f 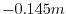 indicates the tank itself.
indicates the tank itself.
- Line 65,
hydrogThetaFile='thetaPol.bin',
This line specifies the name of the file from which the initial values
of temperature
are read. This file is a three-dimensional
(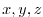 ) map and is enumerated and formatted in the same manner as the
bathymetry file.
) map and is enumerated and formatted in the same manner as the
bathymetry file.
- Lines 66 and 67
tCylIn = 0
tCylOut = 20
These line specify the temperatures in degrees Celsius of the interior
and exterior walls of the tank - typically taken to be icewater on
the inside and room temperature on the outside.
Other lines in the file input/data are standard values
that are described in the MITgcm Getting Started and MITgcm Parameters
notes.
# ====================
# | Model parameters |
# ====================
#
# Continuous equation parameters
&PARM01
tRef=29*20.0,
sRef=29*35.0,
viscAh=5.0E-6,
viscAz=5.0E-6,
no_slip_sides=.FALSE.,
no_slip_bottom=.FALSE.,
diffKhT=2.5E-6,
diffKzT=2.5E-6,
diffKhS=1.0E-6,
diffKzS=1.0E-6,
f0=0.5,
sBeta =0.,
gravity=9.81,
rhoConst=1000.0,
rhoNil=1000.0,
# heatCapacity_Cp=3900.0,
rigidLid=.TRUE.,
implicitFreeSurface=.FALSE.,
eosType='LINEAR',
nonHydrostatic=.TRUE.,
readBinaryPrec=32,
&
# Elliptic solver parameters
&PARM02
cg2dMaxIters=1000,
cg2dTargetResidual=1.E-7,
cg3dMaxIters=10,
cg3dTargetResidual=1.E-9,
&
# Time stepping parameters
&PARM03
nIter0=0,
nTimeSteps=20,
# nTimeSteps=36000000,
deltaT=0.1,
abEps=0.1,
pChkptFreq=1.0,
chkptFreq=1.0,
dumpFreq=1.0,
monitorFreq=0.1,
outputTypesInclusive=.TRUE.,
&
# Gridding parameters
&PARM04
usingCartesianGrid=.FALSE.,
usingCylindricalGrid=.TRUE.,
usingCurvilinearGrid=.FALSE.,
dXspacing=3,
dYspacing=0.01,
delZ=29*0.005,
&
# Input datasets
&PARM05
hydrogThetaFile='thetaPol.bin',
bathyFile='bathyPol.bin',
tCylIn = 0
tCylOut = 20
&
This file uses standard default values and does not contain
customizations for this experiment.
This file uses standard default values and does not contain
customizations for this experiment.
The input/thetaPol.bin file specifies a three-dimensional (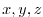 )
map of initial values of
)
map of initial values of 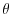 in degrees Celsius. This particular
experiment is set to random values x around 20C to provide initial
perturbations.
in degrees Celsius. This particular
experiment is set to random values x around 20C to provide initial
perturbations.
The input/bathyPol.bin file specifies a two-dimensional (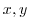 )
map of depth values. For this experiment values are either
)
map of depth values. For this experiment values are either
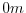 or -delZm, corresponding respectively to outside or inside of
the tank. The file contains a raw binary stream of data that is enumerated
in the same way as standard MITgcm two-dimensional, horizontal arrays.
or -delZm, corresponding respectively to outside or inside of
the tank. The file contains a raw binary stream of data that is enumerated
in the same way as standard MITgcm two-dimensional, horizontal arrays.
Two lines are customized in this file for the current experiment
- Line 39,
sNx=120,
this line sets
the lateral domain extent in grid points for the
axis aligned with the x-coordinate.
- Line 40,
sNy=31,
this line sets
the lateral domain extent in grid points for the
axis aligned with the y-coordinate.
C $Header: /u/gcmpack/manual/s_examples/rotating_tank/code/SIZE.h,v 1.1 2004/07/26 21:09:47 afe Exp $
C $Name: $
C
C /==========================================================\
C | SIZE.h Declare size of underlying computational grid. |
C |==========================================================|
C | The design here support a three-dimensional model grid |
C | with indices I,J and K. The three-dimensional domain |
C | is comprised of nPx*nSx blocks of size sNx along one axis|
C | nPy*nSy blocks of size sNy along another axis and one |
C | block of size Nz along the final axis. |
C | Blocks have overlap regions of size OLx and OLy along the|
C | dimensions that are subdivided. |
C \==========================================================/
C Voodoo numbers controlling data layout.
C sNx - No. X points in sub-grid.
C sNy - No. Y points in sub-grid.
C OLx - Overlap extent in X.
C OLy - Overlat extent in Y.
C nSx - No. sub-grids in X.
C nSy - No. sub-grids in Y.
C nPx - No. of processes to use in X.
C nPy - No. of processes to use in Y.
C Nx - No. points in X for the total domain.
C Ny - No. points in Y for the total domain.
C Nr - No. points in Z for full process domain.
INTEGER sNx
INTEGER sNy
INTEGER OLx
INTEGER OLy
INTEGER nSx
INTEGER nSy
INTEGER nPx
INTEGER nPy
INTEGER Nx
INTEGER Ny
INTEGER Nr
PARAMETER (
& sNx = 120,
& sNy = 31,
& OLx = 3,
& OLy = 3,
& nSx = 1,
& nSy = 1,
& nPx = 1,
& nPy = 1,
& Nx = sNx*nSx*nPx,
& Ny = sNy*nSy*nPy,
& Nr = 29)
C MAX_OLX - Set to the maximum overlap region size of any array
C MAX_OLY that will be exchanged. Controls the sizing of exch
C routine buufers.
INTEGER MAX_OLX
INTEGER MAX_OLY
PARAMETER ( MAX_OLX = OLx,
& MAX_OLY = OLy )
This file uses standard default values and does not contain
customizations for this experiment.
This file uses standard default values and does not contain
customizations for this experiment.



Next: 4. Software Architecture
Up: 3.21 A Rotating Tank
Previous: 3.21.3 Discrete Numerical Configuration
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2018-01-23 |
 |
|







