|
|
|
|



Next: 3.10 Baroclinic Gyre MITgcm
Up: 3.9 Barotropic Gyre MITgcm
Previous: 3.9.2 Discrete Numerical Configuration
Contents
Subsections
3.9.3 Code Configuration
The model configuration for this experiment resides under the
directory verification/tutorial_barotropic_gyre/.
The experiment files
- input/data
- input/data.pkg
- input/eedata,
- input/windx.sin_y,
- input/topog.box,
- code/CPP_EEOPTIONS.h
- code/CPP_OPTIONS.h,
- code/SIZE.h.
contain the code customizations and parameter settings for this
experiments. Below we describe the customizations
to these files associated with this experiment.
This file, reproduced completely below, specifies the main parameters
for the experiment. The parameters that are significant for this configuration
are
- Line 7,
viscAh=4.E2,
this line sets
the Laplacian friction coefficient to
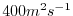
- Line 10,
beta=1.E-11,
this line sets
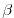 (the gradient of the coriolis parameter,
(the gradient of the coriolis parameter, 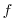 ) to
) to
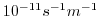
- Lines 15 and 16
rigidLid=.FALSE.,
implicitFreeSurface=.TRUE.,
these lines suppress the rigid lid formulation of the surface
pressure inverter and activate the implicit free surface form
of the pressure inverter.
- Line 27,
startTime=0,
this line indicates that the experiment should start from 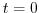 and implicitly suppresses searching for checkpoint files associated
with restarting an numerical integration from a previously saved state.
and implicitly suppresses searching for checkpoint files associated
with restarting an numerical integration from a previously saved state.
- Line 29,
endTime=12000,
this line indicates that the experiment should start finish at 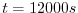 .
A restart file will be written at this time that will enable the
simulation to be continued from this point.
.
A restart file will be written at this time that will enable the
simulation to be continued from this point.
- Line 30,
deltaTmom=1200,
This line sets the momentum equation timestep to 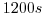 .
.
- Line 39,
usingCartesianGrid=.TRUE.,
This line requests that the simulation be performed in a
Cartesian coordinate system.
- Line 41,
delX=60*20E3,
This line sets the horizontal grid spacing between each x-coordinate line
in the discrete grid. The syntax indicates that the discrete grid
should be comprise of 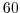 grid lines each separated by
grid lines each separated by
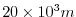 (
(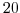 km).
km).
- Line 42,
delY=60*20E3,
This line sets the horizontal grid spacing between each y-coordinate line
in the discrete grid to
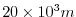 (
(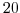 km).
km).
- Line 43,
delZ=5000,
This line sets the vertical grid spacing between each z-coordinate line
in the discrete grid to 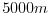 (
(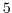 km).
km).
- Line 46,
bathyFile='topog.box'
This line specifies the name of the file from which the domain
bathymetry is read. This file is a two-dimensional (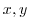 ) map of
depths. This file is assumed to contain 64-bit binary numbers
giving the depth of the model at each grid cell, ordered with the x
coordinate varying fastest. The points are ordered from low coordinate
to high coordinate for both axes. The units and orientation of the
depths in this file are the same as used in the MITgcm code. In this
experiment, a depth of
) map of
depths. This file is assumed to contain 64-bit binary numbers
giving the depth of the model at each grid cell, ordered with the x
coordinate varying fastest. The points are ordered from low coordinate
to high coordinate for both axes. The units and orientation of the
depths in this file are the same as used in the MITgcm code. In this
experiment, a depth of 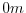 indicates a solid wall and a depth
of
indicates a solid wall and a depth
of 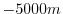 indicates open ocean. The matlab program
input/gendata.m shows an example of how to generate a
bathymetry file.
indicates open ocean. The matlab program
input/gendata.m shows an example of how to generate a
bathymetry file.
- Line 49,
zonalWindFile='windx.sin_y'
This line specifies the name of the file from which the x-direction
surface wind stress is read. This file is also a two-dimensional
( ) map and is enumerated and formatted in the same manner as the
bathymetry file. The matlab program input/gendata.m includes example
code to generate a valid zonalWindFile file.
) map and is enumerated and formatted in the same manner as the
bathymetry file. The matlab program input/gendata.m includes example
code to generate a valid zonalWindFile file.
other lines in the file input/data are standard values
that are described in the MITgcm Getting Started and MITgcm Parameters
notes.
# Model parameters
# Continuous equation parameters
&PARM01
tRef=20.,
sRef=10.,
viscAz=1.E-2,
viscAh=4.E2,
diffKhT=4.E2,
diffKzT=1.E-2,
beta=1.E-11,
tAlpha=2.E-4,
sBeta =0.,
gravity=9.81,
gBaro=9.81,
rigidLid=.FALSE.,
implicitFreeSurface=.TRUE.,
eosType='LINEAR',
readBinaryPrec=64,
&
# Elliptic solver parameters
&PARM02
cg2dMaxIters=1000,
cg2dTargetResidual=1.E-7,
&
# Time stepping parameters
&PARM03
startTime=0,
#endTime=311040000,
endTime=12000.0,
deltaTmom=1200.0,
deltaTtracer=1200.0,
abEps=0.1,
pChkptFreq=2592000.0,
chkptFreq=120000.0,
dumpFreq=2592000.0,
&
# Gridding parameters
&PARM04
usingCartesianGrid=.TRUE.,
usingSphericalPolarGrid=.FALSE.,
delX=60*20E3,
delY=60*20E3,
delZ=5000.,
&
&PARM05
bathyFile='topog.box',
hydrogThetaFile=,
hydrogSaltFile=,
zonalWindFile='windx.sin_y',
meridWindFile=,
&
This file uses standard default values and does not contain
customizations for this experiment.
This file uses standard default values and does not contain
customizations for this experiment.
The input/windx.sin_y file specifies a two-dimensional ( )
map of wind stress ,
)
map of wind stress ,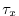 , values. The units used are
, values. The units used are 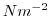 .
Although
.
Although 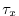 is only a function of
is only a function of 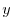 n in this experiment
this file must still define a complete two-dimensional map in order
to be compatible with the standard code for loading forcing fields
in MITgcm. The included matlab program input/gendata.m gives a complete
code for creating the input/windx.sin_y file.
n in this experiment
this file must still define a complete two-dimensional map in order
to be compatible with the standard code for loading forcing fields
in MITgcm. The included matlab program input/gendata.m gives a complete
code for creating the input/windx.sin_y file.
The input/topog.box file specifies a two-dimensional (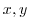 )
map of depth values. For this experiment values are either
)
map of depth values. For this experiment values are either
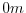 or -delZm, corresponding respectively to a wall or to deep
ocean. The file contains a raw binary stream of data that is enumerated
in the same way as standard MITgcm two-dimensional, horizontal arrays.
The included matlab program input/gendata.m gives a complete
code for creating the input/topog.box file.
or -delZm, corresponding respectively to a wall or to deep
ocean. The file contains a raw binary stream of data that is enumerated
in the same way as standard MITgcm two-dimensional, horizontal arrays.
The included matlab program input/gendata.m gives a complete
code for creating the input/topog.box file.
Two lines are customized in this file for the current experiment
- Line 39,
sNx=60,
this line sets
the lateral domain extent in grid points for the
axis aligned with the x-coordinate.
- Line 40,
sNy=60,
this line sets
the lateral domain extent in grid points for the
axis aligned with the y-coordinate.
C $Header: /u/gcmpack/manual/s_examples/barotropic_gyre/code/SIZE.h,v 1.2 2001/09/27 00:58:17 cnh Exp $
C $Name: $
C
C /==========================================================\
C | SIZE.h Declare size of underlying computational grid. |
C |==========================================================|
C | The design here support a three-dimensional model grid |
C | with indices I,J and K. The three-dimensional domain |
C | is comprised of nPx*nSx blocks of size sNx along one axis|
C | nPy*nSy blocks of size sNy along another axis and one |
C | block of size Nz along the final axis. |
C | Blocks have overlap regions of size OLx and OLy along the|
C | dimensions that are subdivided. |
C \==========================================================/
C Voodoo numbers controlling data layout.
C sNx - No. X points in sub-grid.
C sNy - No. Y points in sub-grid.
C OLx - Overlap extent in X.
C OLy - Overlat extent in Y.
C nSx - No. sub-grids in X.
C nSy - No. sub-grids in Y.
C nPx - No. of processes to use in X.
C nPy - No. of processes to use in Y.
C Nx - No. points in X for the total domain.
C Ny - No. points in Y for the total domain.
C Nr - No. points in R for full process domain.
INTEGER sNx
INTEGER sNy
INTEGER OLx
INTEGER OLy
INTEGER nSx
INTEGER nSy
INTEGER nPx
INTEGER nPy
INTEGER Nx
INTEGER Ny
INTEGER Nr
PARAMETER (
& sNx = 60,
& sNy = 60,
& OLx = 3,
& OLy = 3,
& nSx = 1,
& nSy = 1,
& nPx = 1,
& nPy = 1,
& Nx = sNx*nSx*nPx,
& Ny = sNy*nSy*nPy,
& Nr = 1)
C MAX_OLX - Set to the maximum overlap region size of any array
C MAX_OLY that will be exchanged. Controls the sizing of exch
C routine buufers.
INTEGER MAX_OLX
INTEGER MAX_OLY
PARAMETER ( MAX_OLX = OLx,
& MAX_OLY = OLy )
This file uses standard default values and does not contain
customizations for this experiment.
This file uses standard default values and does not contain
customizations for this experiment.



Next: 3.10 Baroclinic Gyre MITgcm
Up: 3.9 Barotropic Gyre MITgcm
Previous: 3.9.2 Discrete Numerical Configuration
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2018-01-23 |
 |
|







