|
|
|
|



Next: 6.3.3 PTRACERS Package
Up: 6.3 General purpose numerical
Previous: 6.3.1 OBCS: Open boundary
Contents
Subsections
6.3.2 RBCS Package
A package which provides the flexibility
to relax fields (temperature, salinity, ptracers)
in any 3-D location:
so could be used as a sponge layer, or as a
"source" anywhere in the domain.
For a tracer (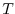 ) at every grid point the tendency is modified so that:
) at every grid point the tendency is modified so that:
where 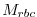 is a 3-D mask (no time dependence) with
values between 0 and 1. Where
is a 3-D mask (no time dependence) with
values between 0 and 1. Where 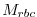 is 1, relaxing timescale
is
is 1, relaxing timescale
is 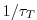 . Where it is 0 there is no relaxing.
The value relaxed to is a 3-D (potentially varying in
time) field given by
. Where it is 0 there is no relaxing.
The value relaxed to is a 3-D (potentially varying in
time) field given by 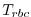 .
.
A seperate mask can be used for T,S and ptracers and
each of these
can be relaxed or not and can have its own timescale
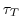 . These are set in data.rbcs (see below).
. These are set in data.rbcs (see below).
The only compile-time parameter you are likely to have to change is in RBCS.h,
the number of masks, PARAMETER(maskLEN = 3 ), see below.
The runtime parameters are set in data.rbcs:
Set in RBCS_PARM01:
 rbcsForcingPeriod: time interval between forcing fields
(in seconds), zero means constant-in-time forcing.
rbcsForcingPeriod: time interval between forcing fields
(in seconds), zero means constant-in-time forcing.
 rbcsForcingCycle: repeat cycle of forcing fields (in seconds),
zero means non-cyclic forcing.
rbcsForcingCycle: repeat cycle of forcing fields (in seconds),
zero means non-cyclic forcing.
 rbcsForcingOffset: time offset of forcing fields
(in seconds, default 0); this is relative to time averages starting at
rbcsForcingOffset: time offset of forcing fields
(in seconds, default 0); this is relative to time averages starting at
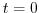 , i.e., the first forcing record/file is placed at
, i.e., the first forcing record/file is placed at
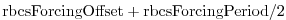 ; see below for examples.
; see below for examples.
 rbcsSingleTimeFiles: true or false (default false),
if true, forcing fields are given 1 file per rbcsForcingPeriod.
rbcsSingleTimeFiles: true or false (default false),
if true, forcing fields are given 1 file per rbcsForcingPeriod.
 deltaTrbcs: time step used to compute the iteration numbers
for rbcsSingleTimeFiles=T.
deltaTrbcs: time step used to compute the iteration numbers
for rbcsSingleTimeFiles=T.
 rbcsIter0: shift in iteration numbers used to label files if
rbcsSingleTimeFiles=T (default 0, see below for examples).
rbcsIter0: shift in iteration numbers used to label files if
rbcsSingleTimeFiles=T (default 0, see below for examples).
 useRBCtemp: true or false (default false)
useRBCtemp: true or false (default false)
 useRBCsalt: true or false (default false)
useRBCsalt: true or false (default false)
 useRBCptracers: true or false (default false), must be using
ptracers to set true
useRBCptracers: true or false (default false), must be using
ptracers to set true
 tauRelaxT: timescale in seconds of relaxing
in temperature (
tauRelaxT: timescale in seconds of relaxing
in temperature (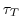 in equation above).
Where mask is 1, relax rate will be
1/tauRelaxT. Default is 1.
in equation above).
Where mask is 1, relax rate will be
1/tauRelaxT. Default is 1.
 tauRelaxS: same for salinity.
tauRelaxS: same for salinity.
 relaxMaskFile(irbc): filename of 3-D file
with mask (
relaxMaskFile(irbc): filename of 3-D file
with mask (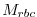 in equation above.
Need a file for each irbc. 1=temperature,
2=salinity, 3=ptracer01, 4=ptracer02 etc. If the mask numbers
end (see maskLEN) are less than the number tracers, then
relaxMaskFile(maskLEN) is used for all remaining ptracers.
in equation above.
Need a file for each irbc. 1=temperature,
2=salinity, 3=ptracer01, 4=ptracer02 etc. If the mask numbers
end (see maskLEN) are less than the number tracers, then
relaxMaskFile(maskLEN) is used for all remaining ptracers.
 relaxTFile: name of file where temperatures
that need to be relaxed to (
relaxTFile: name of file where temperatures
that need to be relaxed to (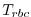 in equation above)
are stored. The file must contain 3-D records to match the model domain.
If rbcsSingleTimeFiles=F, it must have one record for each forcing period.
If T, there must be a separate file for each period and a 10-digit iteration
number is appended to the file name (see Table 6.5
and examples below).
in equation above)
are stored. The file must contain 3-D records to match the model domain.
If rbcsSingleTimeFiles=F, it must have one record for each forcing period.
If T, there must be a separate file for each period and a 10-digit iteration
number is appended to the file name (see Table 6.5
and examples below).
 relaxSFile: same for salinity.
relaxSFile: same for salinity.
Set in RBCS_PARM02 for each of the ptracers (iTrc):
 useRBCptrnum(iTrc): true or false (default
is false).
useRBCptrnum(iTrc): true or false (default
is false).
 tauRelaxPTR(iTrc): relax timescale.
tauRelaxPTR(iTrc): relax timescale.
 relaxPtracerFile(iTrc): file with relax
fields.
relaxPtracerFile(iTrc): file with relax
fields.
For constant-in-time relaxation, set rbcsForcingPeriod=0.
For time-varying relaxation, Table 6.5 illustrates the
relation between model time and forcing fields (either records in
one big file or, for rbcsSingleTimeFiles=T, individual files labeled with an
iteration number). With rbcsSingleTimeFiles=T, this is the same as in the
offline package, except that the forcing offset is in seconds.
Table 6.5:
Timing of relaxation forcing fields.
| |
rbcsSingleTimeFiles = T |
F |
| |
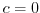
|
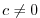
|
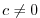
|
| model time |
file number |
file number |
record |
|---|
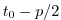
|
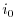
|
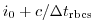
|
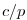
|
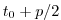
|
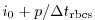
|
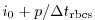
|
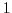
|
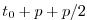
|
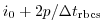
|
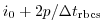
|
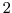
|
| ... |
... |
... |
... |
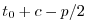
|
... |
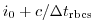
|
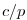
|
| ... |
... |
... |
... |
| |
|
| where |
|
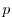
|
$=$ | rbcsForcingPeriod |
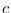
|
$=$ | rbcsForcingCycle |
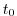
|
$=$ | rbcsForcingOffset |
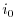
|
$=$ | rbcsIter0 |
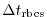
|
$=$ | deltaTrbcs |
|
Set rbcsSingleTimeFiles=F and
rbcsForcingOffset=0, and the model will start by interpolating the last and first
records of rbcs data, placed at 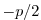 and
and 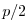 , resp., as appropriate for fields
averaged over the time intervals
, resp., as appropriate for fields
averaged over the time intervals ![$ [-p, 0]$](img2036.png) and
and ![$ [0, p]$](img2037.png) .
.
Set rbcsForcingCycle=0 and
rbcsSingleTimeFiles=T. With rbcsForcingOffset=0, rbcsIter0=0 and
deltaTrbcs=rbcsForcingPeriod, the model would then start by interpolating data from
files relax*File.0000000000.data and relax*File.0000000001.data, ...,
again placed at 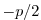 and
and 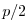 .
.
Set rbcsSingleTimeFiles=F and
rbcsForcingOffset=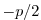 , and the model will start forcing with the first
record at
, and the model will start forcing with the first
record at 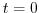 .
.
Set rbcsForcingCycle=0 and
rbcsSingleTimeFiles=T. In this case, it is more natural to set
rbcsForcingOffset=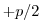 .
With rbcsIter0=0 and deltaTrbcs=rbcsForcingPeriod, the model would then start
with data from files relax*File.0000000000.data at
.
With rbcsIter0=0 and deltaTrbcs=rbcsForcingPeriod, the model would then start
with data from files relax*File.0000000000.data at 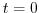 .
It would then proceed to interpolate between this file and files
relax*File.0000000001.data at
.
It would then proceed to interpolate between this file and files
relax*File.0000000001.data at 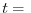 rbcsForcingPeriod.
rbcsForcingPeriod.
6.3.2.8 Experiments and tutorials that use rbcs
In the directory verifcation, the following experiments use
rbcs:



Next: 6.3.3 PTRACERS Package
Up: 6.3 General purpose numerical
Previous: 6.3.1 OBCS: Open boundary
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2018-01-23 |
 |
|







