


Next: 6.3.3 PTRACERS Package
Up: 6.3 General purpose numerical
Previous: 6.3.1 OBCS: Open boundary
Contents
Subsections
6.3.2 RBCS Package
A package which provides the flexibility
to relax fields (temperature, salinity, ptracers)
in any 3-D location:
so could be used as a sponge layer, or as a
"source" anywhere in the domain.
For a tracer (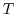 ) at every grid point the tendency is modified so that:
) at every grid point the tendency is modified so that:
where 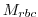 is a 3-D mask (no time dependence) with
values between 0 and 1. Where
is a 3-D mask (no time dependence) with
values between 0 and 1. Where 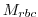 is 1, relaxing timescale
is
is 1, relaxing timescale
is 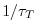 . Where it is 0 there is no relaxing.
The value relaxed to is a 3-D (potentially varying in
time) field given by
. Where it is 0 there is no relaxing.
The value relaxed to is a 3-D (potentially varying in
time) field given by 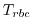 .
.
A seperate mask can be used for T,S and ptracers and
each of these
can be relaxed or not and can have its own timescale
 . These are set in data.rbcs (see below).
. These are set in data.rbcs (see below).
The only change need in the code might be in RBCS.H, for
PARAMETER(maskLEN = 3 ), if you need more than 3
masks (see below).
There are runtime parameters
set in data.rbcs:
These runtime options include
Set in RBCS_PARM01:
 Parameters to set the timing for periodic fields to
relax to are to
be loaded are: rbcs_ForcingPeriod, rbcs_ForcingCycle.
The former is how often to load, the latter is how often to cycle
through those fields (eg. period couple be monthly and cycle one year).
rbcs_ForcingCycle=0 meaning no periodic forcing, and the relax field
is only read in at the beginning of the run and kept constant
the rest of the run. Default is 0.
Parameters to set the timing for periodic fields to
relax to are to
be loaded are: rbcs_ForcingPeriod, rbcs_ForcingCycle.
The former is how often to load, the latter is how often to cycle
through those fields (eg. period couple be monthly and cycle one year).
rbcs_ForcingCycle=0 meaning no periodic forcing, and the relax field
is only read in at the beginning of the run and kept constant
the rest of the run. Default is 0.
 rbcsIniter: if you want to offset rbcs forcing
timing. Default is nIter0.
rbcsIniter: if you want to offset rbcs forcing
timing. Default is nIter0.
 useRBCtemp: true or false (default false)
useRBCtemp: true or false (default false)
 useRBCsalt: true or false (default false)
useRBCsalt: true or false (default false)
 useRBCptracers: true or false (default false), must be using
ptracers to set true
useRBCptracers: true or false (default false), must be using
ptracers to set true
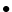 tauRelaxT: timescale in seconds of relaxing
in temperature (
tauRelaxT: timescale in seconds of relaxing
in temperature (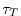 in equation above).
Where mask is 1, relax rate will be
1/tauRelaxT. Default is 1.
in equation above).
Where mask is 1, relax rate will be
1/tauRelaxT. Default is 1.
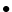 tauRelaxS: same for salinity.
tauRelaxS: same for salinity.
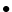 relaxMaskFile(irbc): filename of 3-D file
with mask (
relaxMaskFile(irbc): filename of 3-D file
with mask (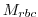 in equation above.
Need a file for each irbc. 1=temperature,
2=salinity, 3=ptracer01, 4=ptracer02 etc. If the mask numbers
end (see maskLEN) are less than the number tracers, then
relaxMaskFile(maskLEN) is used for all remaining ptracers.
in equation above.
Need a file for each irbc. 1=temperature,
2=salinity, 3=ptracer01, 4=ptracer02 etc. If the mask numbers
end (see maskLEN) are less than the number tracers, then
relaxMaskFile(maskLEN) is used for all remaining ptracers.
 relaxTFile: name of file where temperatures
that need to be realxed to (
relaxTFile: name of file where temperatures
that need to be realxed to (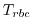 in equation above)
are stored. Need 3-D fields to
match model domain, and as many entries as given by
rbcsForcingPeriod and rbcsForcingCycle.
in equation above)
are stored. Need 3-D fields to
match model domain, and as many entries as given by
rbcsForcingPeriod and rbcsForcingCycle.
 relaxSFile: same for salinity.
relaxSFile: same for salinity.
Set in RBCS_PARM02 for each of the ptracers (iTrc):
 useRBCptrnum(iTrc): true or false (default
is false).
useRBCptrnum(iTrc): true or false (default
is false).
 tauRelaxPTR(iTrc): relax timescale.
tauRelaxPTR(iTrc): relax timescale.
 relaxPtracerFile(iTrc): file with relax
fields.
relaxPtracerFile(iTrc): file with relax
fields.
6.3.2.5 Experiments and tutorials that use rbcs



Next: 6.3.3 PTRACERS Package
Up: 6.3 General purpose numerical
Previous: 6.3.1 OBCS: Open boundary
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2011-01-09 |
 |
|







