|
|
|
|



Next: 7.1.5 Dos and Donts
Up: 7.1 Diagnostics-A Flexible Infrastructure
Previous: 7.1.3 Key Subroutines and
Contents
Subsections
7.1.4 Usage Notes
To use the diagnostics package, other than enabling it in packages.conf
and turning the useDiagnostics flag in data.pkg to .TRUE., there are two
further steps the user must take to enable the diagnostics package for
output of quantities that are already defined in the GCM under an experiment's
configuration of packages.
A parameter file data.diagnostics must be supplied in the run directory,
and the file DIAGNOSTICS_SIZE.h must be included in the
code directory. The steps for defining a new (permanent or experiment-specific
temporary) diagnostic quantity will be outlined later.
The namelist in parameter file data.diagnostics will activate
a user-defined list of diagnostics quantities to be computed,
specify the frequency and type of output, the number of levels, and
the name of all the separate output files.
A sample data.diagnostics namelist file:
# Diagnostic Package Choices
#--------------------
# dumpAtLast (logical): always write output at the end of simulation (default=F)
# diag_mnc (logical): write to NetCDF files (default=useMNC)
#--for each output-stream:
# fileName(n) : prefix of the output file name (max 80c long) for outp.stream n
# frequency(n):< 0 : write snap-shot output every |frequency| seconds
# > 0 : write time-average output every frequency seconds
# timePhase(n) : write at time = timePhase + multiple of |frequency|
# averagingFreq : frequency (in s) for periodic averaging interval
# averagingPhase : phase (in s) for periodic averaging interval
# repeatCycle : number of averaging intervals in 1 cycle
# levels(:,n) : list of levels to write to file (Notes: declared as REAL)
# when this entry is missing, select all common levels of this list
# fields(:,n) : list of selected diagnostics fields (8.c) in outp.stream n
# (see "available_diagnostics.log" file for the full list of diags)
# missing_value(n) : missing value for real-type fields in output file "n"
# fileFlags(n) : specific code (8c string) for output file "n"
#--------------------
&DIAGNOSTICS_LIST
fields(1:2,1) = 'UVEL ','VVEL ',
levels(1:5,1) = 1.,2.,3.,4.,5.,
filename(1) = 'diagout1',
frequency(1) = 86400.,
fields(1:2,2) = 'THETA ','SALT ',
filename(2) = 'diagout2',
fileflags(2) = ' P1 ',
frequency(2) = 3600.,
&
&DIAG_STATIS_PARMS
&
In this example, there are two output files that will be generated for
each tile and for each output time. The first set of output files has
the prefix diagout1, does time averaging every 86400. seconds,
(frequency is 86400.), and will write fields which are multiple-level
fields at output levels 1-5. The names of diagnostics quantities are
UVEL and VVEL. The second set of output files has the prefix
diagout2, does time averaging every 3600. seconds, includes fields
with all levels, and the names of diagnostics quantities are THETA and SALT.
The user must assure that enough computer memory is allocated for the
diagnostics and the output streams selected for a particular
experiment. This is accomplished by modifying the file
DIAGNOSTICS_SIZE.h and including it in the experiment code directory.
The parameters that should be checked are called numDiags, numLists,
numperList, and diagSt_size.
numDiags (and diagSt_size):
All GCM diagnostic quantities are stored in the single diagnostic array QDIAG
which is located in the file
pkg/diagnostics/DIAGNOSTICS.h
and has the form:
_RL qdiag(1-Olx,sNx+Olx,1-Olx,sNx+Olx,numDiags,nSx,nSy)
_RL qSdiag(0:nStats,0:nRegions,diagSt_size,nSx,nSy)
COMMON / DIAG_STORE_R / qdiag, qSdiag
The first two-dimensions of qdiag correspond to the horizontal
dimension of a given diagnostic, and the third dimension of qdiag is
used to identify diagnostic fields and levels combined. In order to
minimize the memory requirement of the model for diagnostics, the
default GCM executable is compiled with room for only one horizontal
diagnostic array, or with numDiags set to Nr. In order for the User to
enable more than 1 three-dimensional diagnostic, the size of the
diagnostics common must be expanded to accommodate the desired
diagnostics. This can be accomplished by manually changing the
parameter numDiags in the file
pkg/diagnostics/DIAGNOSTICS_SIZE.h.
numDiags should be set greater than or equal to the sum of all the
diagnostics activated for output each multiplied by the number of
levels defined for that diagnostic quantity. For the above example,
there are 4 multiple level fields, which the diagnostics menu (see
below) indicates are defined at the GCM vertical resolution, Nr. The
value of numDiags in DIAGNOSTICS_SIZE.h would therefore be equal to
4*Nr, or, say 40 if 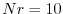 .
.
numLists and numperList:
The parameter numLists must be set greater than or equal to
the number of separate output streams that the user specifies in the
namelist file data.diagnostics. The parameter numperList corresponds
to the maximum number of diagnostics requested per output streams.
In order to define and include as part of the diagnostic output any
field that is desired for a particular experiment, two steps must be
taken. The first is to enable the ``User Diagnostic'' in
data.diagnostics. This is accomplished by adding one of the ``User
Diagnostic'' field names (UDIAG1 through UDIAG10, for multi-level
fields, or SDIAG1 through SDIAG10 for single level fields) to the
data.diagnostics namelist in one of the output streams. These fields
are listed in the diagnostics menu. The second step is to add a call
to DIAGNOSTICS_FILL from the subroutine in which the quantity desired
for diagnostic output is computed.
In order to add a new diagnostic to the permanent set of diagnostics
that the main model or any package contains as part of its diagnostics
menu, the subroutine DIAGNOSTICS_ADDTOLIST should be called during the
initialization phase of the main model or package. For the main model,
the call should be made from subroutine DIAGNOSTICS_MAIN_INIT, and
for a package, the call should probably be made from
from inside the particular package's init_fixed routine.
A typical code sequence to set the
input arguments to DIAGNOSTICS_ADDTOLIST would look like:
diagName = 'RHOAnoma'
diagTitle = 'Density Anomaly (=Rho-rhoConst)'
diagUnits = 'kg/m^3 '
diagCode = 'SMR MR '
CALL DIAGNOSTICS\_ADDTOLIST( diagNum,
I diagName, diagCode, diagUnits, diagTitle, 0, myThid )
If the new diagnostic quantity is associated with either a
vector pair or a diagnostic counter, the diagMate argument must be
provided with the proper index corresponding to the ``mate''.
The output argument from DIAGNOSTICS_ADDTOLIST that is called diagNum here
contains a running total of the number of diagnostics defined in the code up to
any point during the run. The sequence number for the next two
diagnostics defined (the two components of the vector pair, for
instance) will be diagNum+1 and diagNum+2. The definition of the first
component of the vector pair must fill the ``mate'' segment of the
diagCode as diagnostic number diagNum+2. Since the subroutine
increments diagNum, the definition of the second component of the
vector fills the ``mate'' part of diagCode with diagNum. A code
sequence for this case would look like:
diagName = 'UVEL '
diagTitle = 'Zonal Component of Velocity (m/s)'
diagUnits = 'm/s '
diagCode = 'UUR MR '
diagMate = diagNum + 2
CALL DIAGNOSTICS_ADDTOLIST( diagNum,
I diagName, diagCode, diagUnits, diagTitle, diagMate, myThid )
diagName = 'VVEL '
diagTitle = 'Meridional Component of Velocity (m/s)'
diagUnits = 'm/s '
diagCode = 'VVR MR '
diagMate = diagNum
CALL DIAGNOSTICS_ADDTOLIST( diagNum,
I diagName, diagCode, diagUnits, diagTitle, diagMate, myThid )
| NAME |
UNITS |
LEVELS |
DESCRIPTION |
| |
|
|
|
| UVEL |
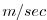
|
Nr |
|
| VVEL |
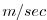
|
Nr |
|
| UVEL_k2 |
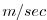
|
1 |
|
| VVEL_k2 |
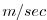
|
1 |
|
| WVEL |
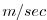
|
Nr |
|
| THETASQ |
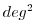
|
Nr |
|
Square of Potential Temperature
|
|
| SALTSQ |
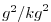
|
Nr |
|
Square of Salt (or Water Vapor Mixing Ratio)
|
|
| SALTSQan |
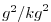
|
Nr |
|
Square of Salt anomaly (=SALT-35)
|
|
| UVELSQ |
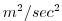
|
Nr |
|
| VVELSQ |
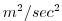
|
Nr |
|
| WVELSQ |
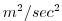
|
Nr |
|
Square of Vertical-Velocity
|
|
| UV_VEL_C |
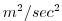
|
Nr |
|
Meridional Transport of Zonal Momentum (cell center)
|
|
| UV_VEL_Z |
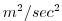
|
Nr |
|
Meridional Transport of Zonal Momentum (corner)
|
|
| WU_VEL |
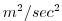
|
Nr |
|
Vertical Transport of Zonal Momentum (cell center)
|
|
| WV_VEL |
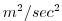
|
Nr |
|
Vertical Transport of Meridional Momentum (cell center)
|
|
| UVELMASS |
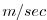
|
Nr |
|
Zonal Mass-Weighted Component of Velocity
|
|
| VVELMASS |
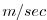
|
Nr |
|
Meridional Mass-Weighted Component of Velocity
|
|
| WVELMASS |
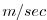
|
Nr |
|
Vertical Mass-Weighted Component of Velocity
|
|
| UTHMASS |
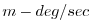
|
Nr |
|
Zonal Mass-Weight Transp of Pot Temp
|
|
| VTHMASS |
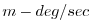
|
Nr |
|
Meridional Mass-Weight Transp of Pot Temp
|
|
| WTHMASS |
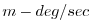
|
Nr |
|
Vertical Mass-Weight Transp of Pot Temp
|
|
| ETAN |
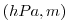
|
1 |
|
Perturbation of Surface (pressure, height)
|
|
| ETANSQ |
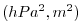
|
1 |
|
Square of Perturbation of Surface (pressure, height)
|
|
| DETADT2 |
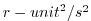
|
1 |
|
Square of Eta (Surf.P,SSH) Tendency
|
|
| THETA |
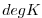
|
Nr |
|
| SST |
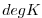
|
1 |
|
| SALT |
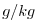
|
Nr |
|
Salt (or Water Vapor Mixing Ratio)
|
|
| SSS |
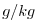
|
1 |
|
| SALTanom |
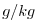
|
Nr |
|
| NAME |
UNITS |
LEVELS |
DESCRIPTION |
| |
|
|
|
| USLTMASS |
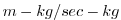
|
Nr |
|
Zonal Mass-Weight Transp of Salt (or W.Vap Mix Rat.)
|
|
| VSLTMASS |
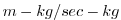
|
Nr |
|
Meridional Mass-Weight Transp of Salt (or W.Vap Mix Rat.)
|
|
| WSLTMASS |
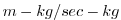
|
Nr |
|
Vertical Mass-Weight Transp of Salt (or W.Vap Mix Rat.)
|
|
| UVELTH |
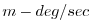
|
Nr |
|
| VVELTH |
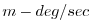
|
Nr |
|
Meridional Transp of Pot Temp
|
|
| WVELTH |
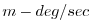
|
Nr |
|
Vertical Transp of Pot Temp
|
|
| UVELSLT |
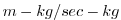
|
Nr |
|
Zonal Transp of Salt (or W.Vap Mix Rat.)
|
|
| VVELSLT |
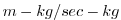
|
Nr |
|
Meridional Transp of Salt (or W.Vap Mix Rat.)
|
|
| WVELSLT |
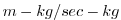
|
Nr |
|
Vertical Transp of Salt (or W.Vap Mix Rat.)
|
|
| RHOAnoma |
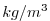
|
Nr |
|
Density Anomaly (=Rho-rhoConst)
|
|
| RHOANOSQ |
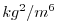
|
Nr |
|
Square of Density Anomaly (=(Rho-rhoConst))
|
|
| URHOMASS |
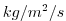
|
Nr |
|
Zonal Transport of Density
|
|
| VRHOMASS |
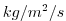
|
Nr |
|
Meridional Transport of Density
|
|
| WRHOMASS |
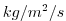
|
Nr |
|
Vertical Transport of Potential Density
|
|
| PHIHYD |
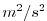
|
Nr |
|
Hydrostatic (ocean) pressure / (atmos) geo-Potential
|
|
| PHIHYDSQ |
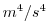
|
Nr |
|
Square of Hyd. (ocean) press / (atmos) geoPotential
|
|
| PHIBOT |
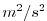
|
Nr |
|
ocean bottom pressure / top. atmos geo-Potential
|
|
| PHIBOTSQ |
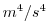
|
Nr |
|
Square of ocean bottom pressure / top. geo-Potential
|
|
| DRHODR |
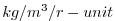
|
Nr |
|
Stratification: d.Sigma/dr
|
|
| VISCA4 |
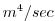
|
1 |
|
Biharmonic Viscosity Coefficient
|
|
| VISCAH |
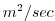
|
1 |
|
Harmonic Viscosity Coefficient
|
|
| TAUX |
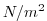
|
1 |
|
zonal surface wind stress, >0 increases uVel
|
|
| TAUY |
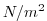
|
1 |
|
meridional surf. wind stress, >0 increases vVel
|
|
| TFLUX |
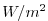
|
1 |
|
net surface heat flux, >0 increases theta
|
|
| TRELAX |
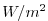
|
1 |
|
surface temperature relaxation, >0 increases theta
|
|
| TICE |
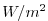
|
1 |
|
heat from melt/freeze of sea-ice, >0 increases theta
|
|
| SFLUX |
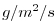
|
1 |
|
net surface salt flux, >0 increases salt
|
|
| SRELAX |
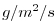
|
1 |
|
surface salinity relaxation, >0 increases salt
|
|
| PRESSURE |
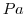
|
Nr |
|
Atmospheric Pressure (Pa)
|
|
| NAME |
UNITS |
LEVELS |
DESCRIPTION |
| |
|
|
|
| ADVr_TH |
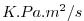
|
Nr |
|
Vertical Advective Flux of Pot.Temperature
|
|
| ADVx_TH |
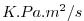
|
Nr |
|
Zonal Advective Flux of Pot.Temperature
|
|
| ADVy_TH |
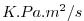
|
Nr |
|
Meridional Advective Flux of Pot.Temperature
|
|
| DFrE_TH |
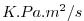
|
Nr |
|
Vertical Diffusive Flux of Pot.Temperature (Explicit part)
|
|
| DIFx_TH |
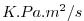
|
Nr |
|
Zonal Diffusive Flux of Pot.Temperature
|
|
| DIFy_TH |
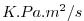
|
Nr |
|
Meridional Diffusive Flux of Pot.Temperature
|
|
| DFrI_TH |
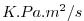
|
Nr |
|
Vertical Diffusive Flux of Pot.Temperature (Implicit part)
|
|
| ADVr_SLT |
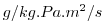
|
Nr |
|
Vertical Advective Flux of Water-Vapor
|
|
| ADVx_SLT |
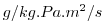
|
Nr |
|
Zonal Advective Flux of Water-Vapor
|
|
| ADVy_SLT |
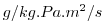
|
Nr |
|
Meridional Advective Flux of Water-Vapor
|
|
| DFrE_SLT |
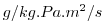
|
Nr |
|
Vertical Diffusive Flux of Water-Vapor (Explicit part)
|
|
| DIFx_SLT |
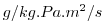
|
Nr |
|
Zonal Diffusive Flux of Water-Vapor
|
|
| DIFy_SLT |
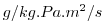
|
Nr |
|
Meridional Diffusive Flux of Water-Vapor
|
|
| DFrI_SLT |
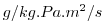
|
Nr |
|
Vertical Diffusive Flux of Water-Vapor (Implicit part)
|
|
| NAME |
UNITS |
LEVELS |
DESCRIPTION |
| |
|
|
|
| SDIAG1 |
|
1 |
|
User-Defined Surface Diagnostic-1
|
|
| SDIAG2 |
|
1 |
|
User-Defined Surface Diagnostic-2
|
|
| SDIAG3 |
|
1 |
|
User-Defined Surface Diagnostic-3
|
|
| SDIAG4 |
|
1 |
|
User-Defined Surface Diagnostic-4
|
|
| SDIAG5 |
|
1 |
|
User-Defined Surface Diagnostic-5
|
|
| SDIAG6 |
|
1 |
|
User-Defined Surface Diagnostic-6
|
|
| SDIAG7 |
|
1 |
|
User-Defined Surface Diagnostic-7
|
|
| SDIAG8 |
|
1 |
|
User-Defined Surface Diagnostic-8
|
|
| SDIAG9 |
|
1 |
|
User-Defined Surface Diagnostic-9
|
|
| SDIAG10 |
|
1 |
|
User-Defined Surface Diagnostic-1-
|
|
| SDIAGC |
|
1 |
|
User-Defined Counted Surface Diagnostic
|
|
| SDIAGCC |
|
1 |
|
User-Defined Counted Surface Diagnostic Counter
|
|
| UDIAG1 |
|
Nrphys |
|
User-Defined Upper-Air Diagnostic-1
|
|
| UDIAG2 |
|
Nrphys |
|
User-Defined Upper-Air Diagnostic-2
|
|
| UDIAG3 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-3
|
|
| UDIAG4 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-4
|
|
| UDIAG5 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-5
|
|
| UDIAG6 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-6
|
|
| UDIAG7 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-7
|
|
| UDIAG8 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-8
|
|
| UDIAG9 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-9
|
|
| UDIAG10 |
|
Nrphys |
|
User-Defined Multi-Level Diagnostic-10
|
|
For a list of the diagnostic fields available in the
different MITgcm packages, follow the link to the diagnostics menu
in the manual section describing the package:



Next: 7.1.5 Dos and Donts
Up: 7.1 Diagnostics-A Flexible Infrastructure
Previous: 7.1.3 Key Subroutines and
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2018-01-23 |
 |
|