|
|
|
|



Next: 3.14 Held-Suarez Atmosphere MITgcm
Up: 3.13 P coordinate Global
Previous: 3.13.2 Discrete Numerical Configuration
Contents
Subsections
3.13.3 Experiment Configuration
The model configuration for this experiment resides under the
directory tutorial_examples/global_ocean_circulation/.
The experiment files
- input/data
- input/data.pkg
- input/eedata,
- input/topog.bin,
- input/deltageopotjmd95.bin,
- input/lev_s.bin,
- input/lev_t.bin,
- input/trenberth_taux.bin,
- input/trenberth_tauy.bin,
- input/lev_sst.bin,
- input/shi_qnet.bin,
- input/shi_empmr.bin,
- code/CPP_EEOPTIONS.h
- code/CPP_OPTIONS.h,
- code/SIZE.h.
contain the code customizations and parameter settings for these
experiments. Below we describe the customizations
to these files associated with this experiment.
Figures (3.5-3.10) show
the relaxation temperature (
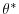 ) and salinity (
) and salinity (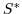 )
fields, the wind stress components (
)
fields, the wind stress components (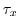 and
and 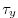 ), the heat
flux (
), the heat
flux (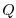 ) and the net fresh water flux (
) and the net fresh water flux (
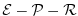 ) used in equations
3.51-3.54.
The figures also indicate the lateral extent and coastline used in the
experiment. Figure (3.11) shows the depth
contours of the model domain.
) used in equations
3.51-3.54.
The figures also indicate the lateral extent and coastline used in the
experiment. Figure (3.11) shows the depth
contours of the model domain.
Figure 3.5:
Annual mean of relaxation temperature [
 ]
]
|
|
Figure 3.6:
Annual mean of relaxation salinity [PSU]
|
|
Figure:
Annual mean of zonal wind stress component [Nmm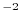 ]
]
|
|
Figure:
Annual mean of meridional wind stress component [Nmm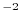 ]
]
|
|
Figure:
Annual mean heat flux [Wm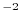 ]
]
|
|
Figure:
Annual mean fresh water flux (Evaporation-Precipitation) [ms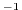 ]
]
|
|
Figure 3.11:
Model bathymetry in pressure units [Pa]
|
|
This file, reproduced completely below, specifies the main parameters
for the experiment. The parameters that are significant for this configuration
are
- Line 15,
viscAr=1.721611620915750E+05,
this line sets the vertical Laplacian dissipation coefficient to
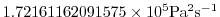 . Note that, the factor
. Note that, the factor
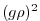 needs to be included in this line. Boundary conditions
for this operator are specified later. This variable is copied into
model general vertical coordinate variable viscAr.
needs to be included in this line. Boundary conditions
for this operator are specified later. This variable is copied into
model general vertical coordinate variable viscAr.

- Line 9-10,
viscAh=3.E5,
no_slip_sides=.TRUE.
these lines set the horizontal Laplacian frictional dissipation
coefficient to
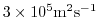 and specify a
no-slip boundary condition for this operator, that is,
and specify a
no-slip boundary condition for this operator, that is, 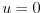 along
boundaries in
along
boundaries in 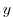 and
and 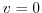 along boundaries in
along boundaries in 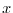 .
.
- Lines 11-13,
viscAr =1.721611620915750e5,
#viscAz =1.67E-3,
no_slip_bottom=.FALSE.,
These lines set the vertical Laplacian frictional dissipation
coefficient to
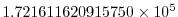 Pa Pa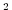 s
s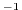 , which corresponds to
, which corresponds to
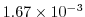 m m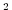 s
s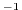 in the commented line, and
specify a free slip boundary condition for this operator, that is,
in the commented line, and
specify a free slip boundary condition for this operator, that is,
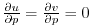 at
at
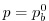 , where
, where 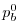 is the local bottom pressure of the
domain at rest. Note that, the factor
is the local bottom pressure of the
domain at rest. Note that, the factor 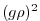 needs to be
included in this line.
needs to be
included in this line.
- Line 14,
diffKhT=1.E3,
this line sets the horizontal diffusion coefficient for temperature
to
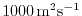 . The boundary condition on this
operator is
. The boundary condition on this
operator is
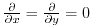 on all boundaries.
on all boundaries.
- Line 15-16,
diffKrT=5.154525811125000e3,
#diffKzT=0.5E-4,
this line sets the vertical diffusion coefficient for temperature to
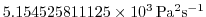 , which
corresponds to
, which
corresponds to
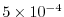 m m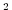 s
s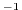 in the commented
line. Note that, the factor
in the commented
line. Note that, the factor 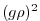 needs to be included in this
line. The boundary condition on this operator is
needs to be included in this
line. The boundary condition on this operator is
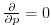 at both the upper and lower
boundaries.
at both the upper and lower
boundaries.
- Line 17-19,
diffKhS=1.E3,
diffKrS=5.154525811125000e3,
#diffKzS=0.5E-4,
These lines set the same values for the diffusion coefficients for
salinity as for temperature.
- Line 20-22,
implicitDiffusion=.TRUE.,
ivdc_kappa=1.030905162225000E9,
#ivdc_kappa=10.0,
Select implicit diffusion as a convection scheme and set coefficient
for implicit vertical diffusion to
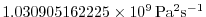 , which corresponds to
, which corresponds to
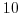 m m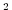 s
s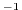 .
.
- Line 23-24,
gravity=9.81,
gravitySign=-1.D0,
These lines set the gravitational acceleration coefficient to
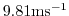 and define the upward direction relative
to the direction of increasing vertical coordinate (in pressure
coordinates, up is in the direction of decreasing pressure)
and define the upward direction relative
to the direction of increasing vertical coordinate (in pressure
coordinates, up is in the direction of decreasing pressure)
- Line 25,
rhoNil=1035.,
sets the reference density of sea water to
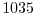 kg m kg m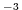 .
.
- Line 28
eosType='JMD95P',
Selects the full equation of state according to Jackett and McDougall
Jackett and McDougall [1995]. The only other sensible choice is the equation of
state by McDougall et al. McDougall et al. [2003], 'MDJFW'. All other
equations of state do not make sense in this configuration.

- Line 28-29,
rigidLid=.FALSE.,
implicitFreeSurface=.TRUE.,
Selects the barotropic pressure equation to be the implicit free
surface formulation.
- Line 30
exactConserv=.TRUE.,
Select a more accurate conservation of properties at the surface
layer by including the horizontal velocity divergence to update the
free surface.
- Line 31-33
nonlinFreeSurf=3,
hFacInf=0.2,
hFacSup=2.0,
Select the nonlinear free surface formulation and set lower and
upper limits for the free surface excursions.
- Line 34
useRealFreshWaterFlux=.FALSE.,
Select virtual salt flux boundary condition for salinity. The
freshwater flux at the surface only affect the surface salinity, but
has no mass flux associated with it
- Line 35-36,
readBinaryPrec=64,
writeBinaryPrec=64,
Sets format for reading binary input datasets and
writing binary output datasets holding model fields to
use 64-bit representation for floating-point numbers.
- Line 42,
cg2dMaxIters=200,
Sets maximum number of iterations the two-dimensional, conjugate
gradient solver will use, irrespective of convergence
criteria being met.

- Line 43,
cg2dTargetResidual=1.E-13,
Sets the tolerance which the two-dimensional, conjugate
gradient solver will use to test for convergence in equation
2.20 to
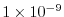 .
Solver will iterate until tolerance falls below this value or until the
maximum number of solver iterations is reached.
.
Solver will iterate until tolerance falls below this value or until the
maximum number of solver iterations is reached.

- Line 48,
startTime=0,
Sets the starting time for the model internal time counter.
When set to non-zero this option implicitly requests a
checkpoint file be read for initial state.
By default the checkpoint file is named according to
the integer number of time steps in the startTime value.
The internal time counter works in seconds.
- Line 49-50,
endTime=8640000.,
#endTime=62208000000,
Sets the time (in seconds) at which this simulation will terminate.
At the end of a simulation a checkpoint file is automatically
written so that a numerical experiment can consist of multiple
stages. The commented out setting for endTime is for a 2000 year
simulation.
- Line 51-53,
deltaTmom = 1200.0,
deltaTtracer = 172800.0,
deltaTfreesurf = 172800.0,
Sets the timestep
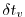 used in the momentum equations to
used in the momentum equations to
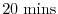 and the timesteps
and the timesteps
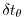 in the tracer
equations and
in the tracer
equations and
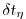 in the implicit free surface
equation to
in the implicit free surface
equation to
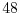 hours
.
See section 2.2. hours
.
See section 2.2.

- Line 55,
pChkptFreq =3110400000.,
write a pick-up file every 100 years of integration.
- Line 56-58
dumpFreq = 3110400000.,
taveFreq = 3110400000.,
monitorFreq = 31104000.,
write model output and time-averaged model output every 100 years,
and monitor statisitics every year.
- Line 59-61
periodicExternalForcing=.TRUE.,
externForcingPeriod=2592000.,
externForcingCycle=31104000.,
Allow periodic external forcing, set forcing period, during which
one set of data is valid, to 1 month and the repeat cycle to 1 year.

- Line 62
tauThetaClimRelax=5184000.0,
Set the restoring timescale to 2 months.

- Line 63
abEps=0.1,
Adams-Bashford factor (see section 2.5)
- Line 68-69
usingCartesianGrid=.FALSE.,
usingSphericalPolarGrid=.TRUE.,
Select spherical grid.
- Line 70-71
dXspacing=4.,
dYspacing=4.,
Set the horizontal grid spacing in degrees spherical distance.
- Line 72
Ro_SeaLevel=53023122.566084,
specifies the total height (in 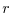 -units, i.e., pressure units) of the
sea surface at rest. This is a reference value.
-units, i.e., pressure units) of the
sea surface at rest. This is a reference value.
- Line 73
groundAtK1=.TRUE.,
specifies the reversal of the vertical indexing. The vertical index is
1 at the bottom of the doman and maximal (i.e., 15) at the surface.
- Line 74-78
delR=7103300.720021, \ldots
set the layer thickness in pressure units, starting with the bottom
layer.
- Line 84-93,
bathyFile='topog.box'
ploadFile='deltageopotjmd95.bin'
hydrogThetaFile='lev_t.bin',
hydrogSaltFile ='lev_s.bin',
zonalWindFile ='trenberth_taux.bin',
meridWindFile ='trenberth_tauy.bin',
thetaClimFile ='lev_sst.bin',
surfQFile ='shi_qnet.bin',
EmPmRFile ='shi_empmr.bin',
This line specifies the names of the files holding the bathymetry
data set, the
time-independent geopotential height anomaly at the bottom, initial
conditions of temperature and salinity, wind stress forcing fields,
sea surface temperature climatology, heat flux, and fresh water flux
(evaporation minus precipitation minus run-off) at the surface.
See file descriptions in section 3.13.3.
other lines in the file input/data are standard values
that are described in the MITgcm Getting Started and MITgcm Parameters
notes.
# ====================
# | Model parameters |
# ====================
#
# Continuous equation parameters
&PARM01
tRef=15*20.,
sRef=15*35.,
viscAh =3.E5,
no_slip_sides=.TRUE.,
viscAr =1.721611620915750e5,
#viscAz =1.67E-3,
no_slip_bottom=.FALSE.,
diffKhT=1.E3,
diffKrT=5.154525811125000e3,
#diffKzT=0.5E-4,
diffKhS=1.E3,
diffKrS=5.154525811125000e3,
#diffKzS=0.5E-4,
implicitDiffusion=.TRUE.,
ivdc_kappa=1.030905162225000e9,
#ivdc_kappa=10.0,
gravity=9.81,
gravitySign=-1.D0,
rhonil=1035.,
buoyancyRelation='OCEANICP',
eosType='JMD95P',
rigidLid=.FALSE.,
implicitFreeSurface=.TRUE.,
exactConserv=.TRUE.,
nonlinFreeSurf=3,
hFacInf=0.2,
hFacSup=2.0,
useRealFreshWaterFlux=.FALSE.,
readBinaryPrec=64,
writeBinaryPrec=64,
cosPower=0.5,
&
# Elliptic solver parameters
&PARM02
cg2dMaxIters=200,
cg2dTargetResidual=1.E-9,
&
# Time stepping parameters
&PARM03
startTime = 0.,
endTime = 8640000.,
#endTime = 62208000000.,
deltaTmom = 1200.0,
deltaTtracer = 172800.0,
deltaTfreesurf = 172800.0,
deltaTClock = 172800.0,
pChkptFreq = 3110400000.,
dumpFreq = 3110400000.,
taveFreq = 3110400000.,
monitorFreq = 31104000.,
periodicExternalForcing=.TRUE.,
externForcingPeriod=2592000.,
externForcingCycle=31104000.,
tauThetaClimRelax=5184000.0,
abEps=0.1,
&
# Gridding parameters
&PARM04
usingCartesianGrid=.FALSE.,
usingSphericalPolarGrid=.TRUE.,
dXspacing=4.,
dYspacing=4.,
Ro_SeaLevel=53023122.566084,
groundAtK1=.TRUE.,
delR=7103300.720021, 6570548.440790, 6041670.010249,
5516436.666057, 4994602.034410, 4475903.435290,
3960063.245801, 3446790.312651, 2935781.405664,
2426722.705046, 1919291.315988, 1413156.804970,
1008846.750166, 705919.025481, 504089.693499,
phiMin=-80.,
&
# Input datasets
&PARM05
topoFile ='topog.bin',
pLoadFile ='deltageopotjmd95.bin',
hydrogThetaFile='lev_t.bin',
hydrogSaltFile ='lev_s.bin',
zonalWindFile ='trenberth_taux.bin',
meridWindFile ='trenberth_tauy.bin',
thetaClimFile ='lev_sst.bin',
#saltClimFile ='lev_sss.bin',
surfQFile ='shi_qnet.bin',
EmPmRFile ='shi_empmr.bin',
&
This file uses standard default values and does not contain
customisations for this experiment.
This file uses standard default values and does not contain
customisations for this experiment.
This file is a two-dimensional (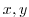 ) map of
depths. This file is assumed to contain 64-bit binary numbers giving
the depth of the model at each grid cell, ordered with the x
coordinate varying fastest. The points are ordered from low
coordinate to high coordinate for both axes. The units and
orientation of the depths in this file are the same as used in the
MITgcm code (Pa for this experiment). In this experiment, a depth of
0 Pa
indicates a land point wall and a depth of
) map of
depths. This file is assumed to contain 64-bit binary numbers giving
the depth of the model at each grid cell, ordered with the x
coordinate varying fastest. The points are ordered from low
coordinate to high coordinate for both axes. The units and
orientation of the depths in this file are the same as used in the
MITgcm code (Pa for this experiment). In this experiment, a depth of
0 Pa
indicates a land point wall and a depth of
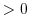 Pa
indicates open ocean. Pa
indicates open ocean.
The file contains 12 identical two dimensional maps (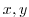 ) of
geopotential height anomaly at the bottom at rest. The values have
been obtained by vertically integrating the hydrostatic equation with
the initial density field (from input/lev_t/s.bin). This file
has to be consitent with the temperature and salinity field at rest
and the choice of equation of state!
) of
geopotential height anomaly at the bottom at rest. The values have
been obtained by vertically integrating the hydrostatic equation with
the initial density field (from input/lev_t/s.bin). This file
has to be consitent with the temperature and salinity field at rest
and the choice of equation of state!
The files input/lev_t/s.bin specify the initial conditions for
temperature and salinity for every grid point in a three dimensional
array (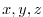 ). The data are obtain by interpolating Levitus
Levitus and T.P.Boyer [1994b] monthly mean values for January onto the model
grid. Keep in mind, that the first index corresponds to the bottom
layer and highest index to the surface layer.
). The data are obtain by interpolating Levitus
Levitus and T.P.Boyer [1994b] monthly mean values for January onto the model
grid. Keep in mind, that the first index corresponds to the bottom
layer and highest index to the surface layer.
Each of the input/trenberth_taux/y.bin files specifies 12
two-dimensional (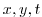 ) maps of zonal and meridional wind stress
values,
) maps of zonal and meridional wind stress
values, 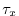 and
and 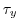 , that is monthly mean values from
Trenberth Trenberth et al. [1990]. The units used are
, that is monthly mean values from
Trenberth Trenberth et al. [1990]. The units used are 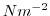 .
.
The file input/lev_sst.bin contains 12 monthly surface
temperature climatologies from Levitus Levitus and T.P.Boyer [1994b] in a three
dimensional array (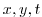 ).
).
The files input/shi_qnet/empmr.bin contain 12 monthly surface
fluxes of heat (qnet) and freshwater (empmr) by Jiang et al.
Jiang et al. [1999] in three dimensional arrays (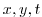 ). Both fluxes are
normalized so that of one year there is no net flux into the
ocean. The freshwater flux is actually constant in time.
). Both fluxes are
normalized so that of one year there is no net flux into the
ocean. The freshwater flux is actually constant in time.
Three lines are customized in this file for the current experiment
- Line 39,
sNx=90,
this line sets
the lateral domain extent in grid points for the
axis aligned with the x-coordinate.
- Line 40,
sNy=40,
this line sets
the lateral domain extent in grid points for the
axis aligned with the y-coordinate.
- Line 49,
Nr=15,
this line sets
the vertical domain extent in grid points.
C $Header: /u/gcmpack/manual/s_examples/global_oce_in_p/code/SIZE.h,v 1.1 2002/12/17 14:39:53 mlosch Exp $
C $Name: $
C
C /==========================================================\
C | SIZE.h Declare size of underlying computational grid. |
C |==========================================================|
C | The design here support a three-dimensional model grid |
C | with indices I,J and K. The three-dimensional domain |
C | is comprised of nPx*nSx blocks of size sNx along one axis|
C | nPy*nSy blocks of size sNy along another axis and one |
C | block of size Nz along the final axis. |
C | Blocks have overlap regions of size OLx and OLy along the|
C | dimensions that are subdivided. |
C \==========================================================/
C Voodoo numbers controlling data layout.
C sNx - No. X points in sub-grid.
C sNy - No. Y points in sub-grid.
C OLx - Overlap extent in X.
C OLy - Overlat extent in Y.
C nSx - No. sub-grids in X.
C nSy - No. sub-grids in Y.
C nPx - No. of processes to use in X.
C nPy - No. of processes to use in Y.
C Nx - No. points in X for the total domain.
C Ny - No. points in Y for the total domain.
C Nr - No. points in Z for full process domain.
INTEGER sNx
INTEGER sNy
INTEGER OLx
INTEGER OLy
INTEGER nSx
INTEGER nSy
INTEGER nPx
INTEGER nPy
INTEGER Nx
INTEGER Ny
INTEGER Nr
PARAMETER (
& sNx = 90,
& sNy = 40,
& OLx = 3,
& OLy = 3,
& nSx = 1,
& nSy = 1,
& nPx = 1,
& nPy = 1,
& Nx = sNx*nSx*nPx,
& Ny = sNy*nSy*nPy,
& Nr = 15)
C MAX_OLX - Set to the maximum overlap region size of any array
C MAX_OLY that will be exchanged. Controls the sizing of exch
C routine buufers.
INTEGER MAX_OLX
INTEGER MAX_OLY
PARAMETER ( MAX_OLX = OLx,
& MAX_OLY = OLy )
This file uses mostly standard default values except for:
#define ATMOSPHERIC_LOADING
enable pressure loading which is abused to include the initial
geopotential height anomaly
#define EXACT_CONSERV
enable more accurate conservation properties to include the
horizontal mass divergence in the free surface
#define NONLIN_FRSURF
enable the nonlinear free surface
This file uses standard default values and does not contain
customisations for this experiment.



Next: 3.14 Held-Suarez Atmosphere MITgcm
Up: 3.13 P coordinate Global
Previous: 3.13.2 Discrete Numerical Configuration
Contents
mitgcm-support@mitgcm.org
| Copyright © 2006
Massachusetts Institute of Technology |
Last update 2011-01-09 |
 |
|







